Enzymes: Isozymes and Regulation
Isozymes:
>Catalyze the same reaction but their physical and chemical properties exhibit significant differences.
>Physically distinct and separable (electrophoresis) forms of a given enzyme present in different cell types.
Nonfunctional Plasma Enzymes:
>Enzymes with no physiologic function in blood whether or not a substrate of the enzyme is present.
>Co-factors are lacking or too dilute.
>The enzyme concentration is far too low.
>The appearance of such enzymes in the plasma reflects destruction of cells from which these enzymes originate.
>This is based upon the assumption that organs contain a specific complement of enzymes and that when the organ is damaged cells in the organ lyse.
Principal Serum Enzymes Used in Clinical Diagnosis:
| Serum Enzyme | Major Diagnostic Use |
| Acid Phosphatase | Carcinoma of the prostate |
| Alanine Aminotransferase (ALT) | Hepatitis |
| Alkaline Phosphatase | Bone disorders; Obstructive liver disease |
| Asparate Aminotransferase (AST) | Myocardial infarction |
| Amylase | Acute pancreatitis |
| Ceruloplasmin | Wilson's disease |
| Creatine Phosphokinase | Myocardial infarction |
| Glutamyl Transpeptidase | Liver disease |
| Lactate Dehydrogenase (LDH) | Myocardial infarction |
| Troponin | Myocardial infarction |
| Lipase | Acute pancreatitis |
Knowledge about the location of different enzymes allows a physician to determine the source of disease.
Three Important Isozymes:
Creatine Phosphokinase (CPK): Creatine + ATP -----> Phosphocreatine + ADP
Tissue specific isozyme, dimer consisting of two subunits, M and B
MM isozyme - muscle
BB isozyme - brain
MB isozyme - heart
Lactate Dehydrogenase (LDH): Pyruvate + NADH -----> Lactate + NAD+
Tissue specific isozyme, tetramer consisting of two subunits, H and M
H4 isozyme - heart (aerobic, requires oxygen)
M4 isozyme - muscle (anaerobic, don't require oxygen)
Troponin: T and I are cardiac regulatory proteins that control the calcium mediated interaction between actin and myosin.
Amylase: Starch -----> "Glucose"
Tissue specific isozyme, monomer
Salivary- occurs in all other tissues.
Pancreatic- absence of this isozyme is indicative of pancreatic dysfunction.
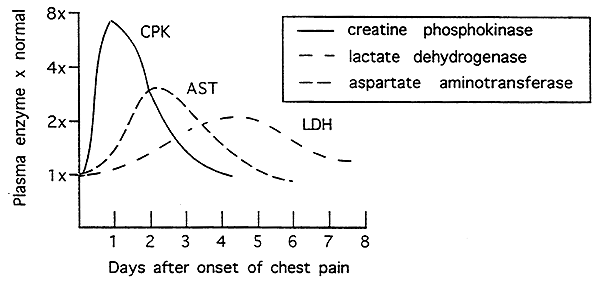
Clinical Correlate: Myocardial Infarction (MI):
Coronary arteries become clogged blocking blood flow to the heart (cardiac muscle).
Some cells do not survive this period of no blood flow.
Following a heart attack the cardiac cells lyse spilling their contents into the blood.
Creatine Phosphokinase (CPK)- MB isozyme appears w/in 24 hrs., normal w/in 48 hrs.
Lactate Dehydrogenase (LDH)- H4 isozyme may persist for two weeks
Aspartate Amino Transferase (AST)- peaks at 2 days, returns to normal by 4 days. (mitochondrial/cytoplasmic isozyme)
Cardiac Troponins (T and I)- appears first w/in 4-6 hrs., may be high for up to 2 weeks

Regulation of Enzyme Activity:
There are five primary forms of enzyme regulation: substrate availability, allosteric, post-translational modification, interaction with control proteins, zymogens.1. Substrate Availability:
Activity determined solely by the concentration (availability) of substrate.
2. Allosteric Regulation:
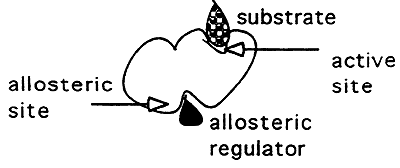
Binding of a non-competitive inhibitor.
Inhibitor binds to a site other than the active site -----> (-) cooperativity, this binding inhibits binding at the active site.
Active Enzyme- Relaxed state (high affinity for substrate)
Inactive Enzyme- Tense state (low affinity for substrate)
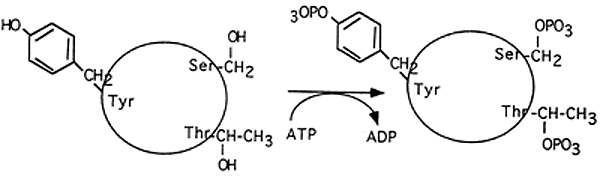
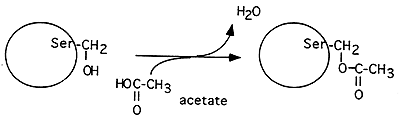
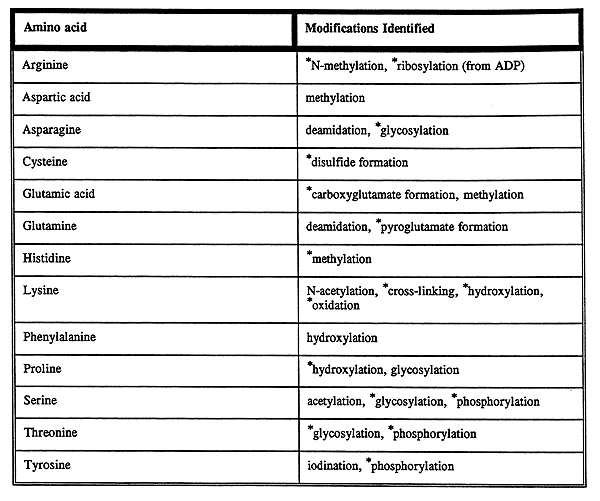
3. Post-translational Modification:
Chemical change in a protein by the covalent attachment of some chemical group to an amino acid.
Phosphorylation (Ser, Thr, Tyr)
Acylation (attachment of -O=C-CH3)
Hydroxylation (Pro and Lys)
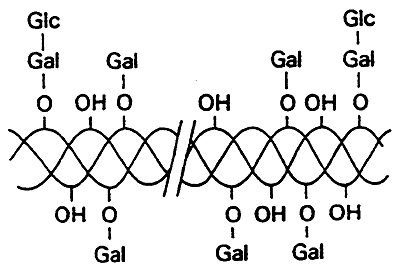
Glycosylation (attachment of sugars)
ADP-Ribosylation (will discuss later)
Carboxylation (attachment of -O2C)
More types of post-translational modification:
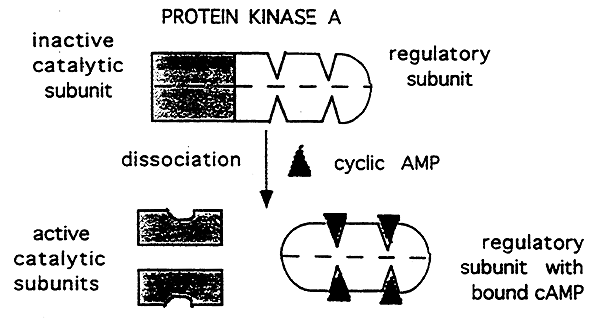
4. Interactions with Control proteins:
Attachment of an inhibitory subunit.
5. Zymogens:
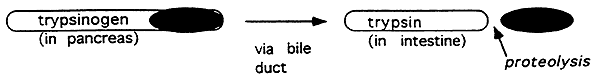
Inactive large precursors of the active form of a protein. The enzymes are activated by removal of a portion of the peptide chain (proteolysis).
Enzyme Mechanisms:
There are three well known enzymes that go through the serine protease mechanism of action, they are: chymotrypsin, trypsin and elastase. We will look at the enzyme mechanism of chymotrypsin in detail.
A protease is an enzyme that hydrolyzes peptide bonds that link amino acids together in a protein. Proteases are specific for certain amino acids and can hydrolyze those amino acids on the carboxy or amino side of the peptide bond.
There are two general types of proteases:
Endoproteases (Serine Proteases):
Can cleave specific peptide bonds within the protein.
Exoproteases:
Cleave only terminal amino acid residues.
Proenzymes (Zymogens):
An enzyme synthesized initially in an inactive form, but are present and poised to act quickly when needed. Serine Proteases are good examples of proenzymes or zymogens.
Physiological Roles for Proenzymes such as Serine Proteases:
1) Digestion of proteins in the small intestine.
i.e. Trypsinogen (proenzyme) ---------> Trypsin (active form)
2) Blood Coagulation
Chymotrypsin:
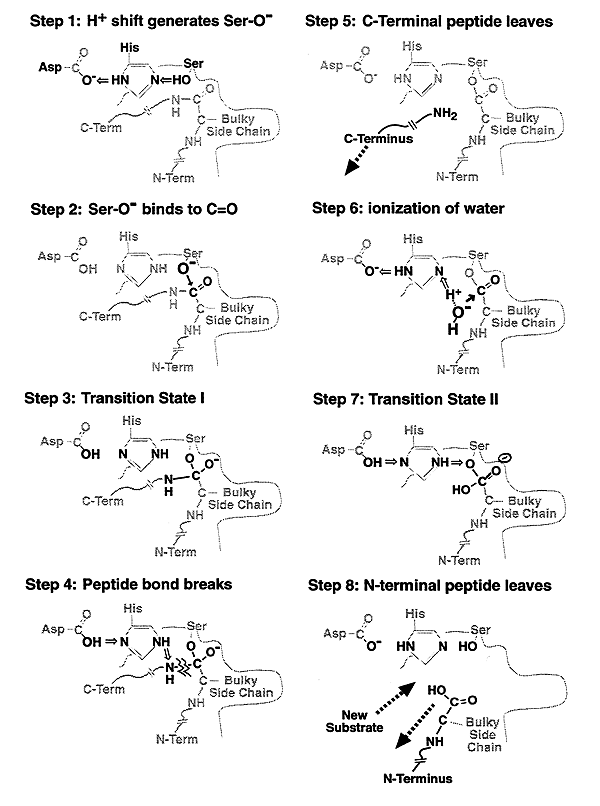
>Used as an example of a serine protease because it's structure and mechanism are well understood.
>Catalyzes the hydrolysis of peptide bonds, on the carboxyl side of bulky aromatic side chains (Tyr, Phe, Trp).
Active Site:
1) Serine, to which the substrate binds, all serine protease active sites contain serine.
2) Histidine, ability to donate and accept protons.
3) Aspartate, ability to accept protons, final link necessary for chymotrypsin action.
Notes:
>These residues are polar (hydrophilic) so would not ordinarily be found on the "interior" of a protein and would normally be deprotonated at physiological pH, except for their critical role in catalysis.
>Though they are in close proximity in the 30 structure, they are not adjacent in the primary sequence (Ser-195, His-57, Asp-102).
Proton Shuttle:
"catalytic triad"
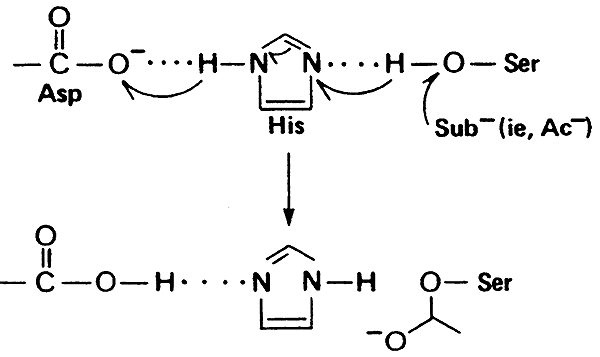
Mechanism of Chymotrypsin Action: "Nucleophilic Acyl Substitution" and "Hydroxylation"
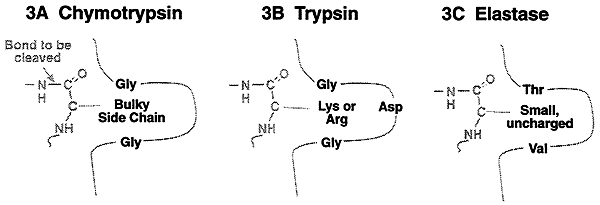
Other Serine Proteases:
>Share the same mechanism of catalysis, but differ in their binding site architecture are trypsin and elastase. Trypsin is specific for the basic amino acids, lysine and arginine. Elastase is specific for the small uncharged amino acids like alanine, valine, threonine etc.
© Dr. Noel Sturm 2021