DNA Mutation and Repair
A mutation, which may arise during replication and/or recombination, is a permanent change in the nucleotide sequence of DNA. Damaged DNA can be mutated either by substitution, deletion or insertion of base pairs. Mutations, for the most part, are harmless except when they lead to cell death or tumor formation. Because of the lethal potential of DNA mutations cells have evolved mechanisms for repairing damaged DNA.
Types of Mutations
There are three types of DNA Mutations: base substitutions, deletions and insertions.
1. Base Substitutions
Single base substitutions are called point mutations, recall the point mutation Glu -----> Val which causes sickle-cell disease. Point mutations are the most common type of mutation and there are two types.
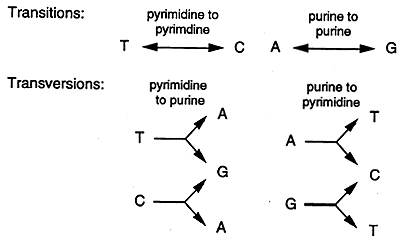
Transition: this occurs when a purine is substituted with another purine or when a pyrimidine is substituted with another pyrimidine.
Transversion: when a purine is substituted for a pyrimidine or a pyrimidine replaces a purine.

Point mutations that occur in DNA sequences encoding proteins are either silent, missense or nonsense.
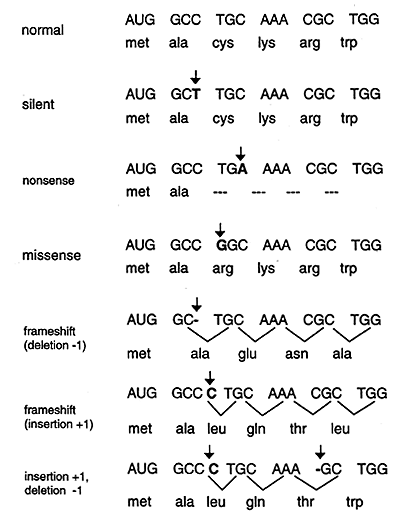
Silent: If abase substitution occurs in the third position of the codon there is a good chance that a synonymous codon will be generated. Thus the amino acid sequence encoded by the gene is not changed and the mutation is said to be silent.
Missence: When base substitution results in the generation of a codon that specifies a different amino acid and hence leads to a different polypeptide sequence. Depending on the type of amino acid substitution the missense mutation is either conservative or nonconservative. For example if the structure and properties of the substituted amino acid are very similar to the original amino acid the mutation is said to be conservative and will most likely have little effect on the resultant proteins structure / function. If the substitution leads to an amino acid with very different structure and properties the mutation is nonconservative and will probably be deleterious (bad) for the resultant proteins structure / function (i.e. the sickle cell point mutation).
Nonsense: When a base substitution results in a stop codon ultimately truncating translation and most likely leading to a nonfunctional protein.
2. Deletions
A deletion, resulting in a frameshift, results when one or more base pairs are lost from the DNA (see Figure above). If one or two bases are deleted the translational frame is altered resulting in a garbled message and nonfunctional product. A deletion of three or more bases leave the reading frame intact. A deletion of one or more codons results in a protein missing one or more amino acids. This may be deleterious or not.
3. Insertions
The insertion of additional base pairs may lead to frameshifts depending on whether or not multiples of three base pairs are inserted. Combinations of insertions and deletions leading to a variety of outcomes are also possible.
Causes of Mutations
Errors in DNA Replication
On very, very rare occasions DNA polymerase will incorporate a noncomplementary base into the daughter strand. During the next round of replication the missincorporated base would lead to a mutation. This, however, is very rare as the exonuclease functions as a proofreading mechanism recognizing mismatched base pairs and excising them.
Errors in DNA Recombination
DNA often rearranges itself by a process called recombination which proceeds via a variety of mechanisms. Occasionally DNA is lost during replication leading to a mutation.
Chemical Damage to DNA
Many chemical mutagens, some exogenous, some man-made, some environmental, are capable of damaging DNA. Many chemotherapeutic drugs and intercalating agent drugs function by damaging DNA.
Radiation
Gamma rays, X-rays, even UV light can interact with compounds in the cell generating free radicals which cause chemical damage to DNA.
DNA Repair
Damaged DNA can be repaired by several different mechanisms.
Mismatch Repair
Sometimes DNA polymerase incorporates an incorrect nucleotide during strand synthesis and the 3' to 5' editing system, exonuclease, fails to correct it. These mismatches as well as single base insertions and deletions are repaired by the mismatch repair mechanism. Mismatch repair relies on a secondary signal within the DNA to distinguish between the parental strand and daughter strand, which contains the replication error. Human cells posses a mismatch repair system similar to that of E. coli, which is described here. Methylation of the sequence GATC occurs on both strands sometime after DNA replication. Because DNA replication is semi-conservative, the new daughter strand remains unmethylated for a very short period of time following replication. This difference allows the mismatch repair system to determine which strand contains the error. A protein, MutS recognizes and binds the mismatched base pair.
Another protein, MutL then binds to MutS and the partially methylated GATC sequence is recognized and bound by the endonuclease, MutH. The MutL/MutS complex then links with MutH which cuts the unmethylated DNA strand at the GATC site. A DNA Helicase, MutU unwinds the DNA strand in the direction of the mismatch and an exonuclease degrades the strand. DNA polymerase then fills in the gap and ligase seals the nick. Defects in the mismatch repair genes found in humans appear to be associated with the development of hereditary colorectal cancer.
Nucleotide Excision Repair (NER)
NER in human cells begins with the formation of a complex of proteins XPA, XPF, ERCC1, HSSB at the lesion on the DNA. The transcription factor TFIIH, which contains several proteins, then binds to the complex in an ATP dependent reaction and makes an incision. The resulting 29 nucleotide segment of damaged DNA is then unwound, the gap is filled (DNA polymerase) and the nick sealed (ligase).
Direct Repair of Damaged DNA
Sometimes damage to a base can be directly repaired by specialized enzymes without having to excise the nucleotide.
Recombination Repair
This mechanism enables a cell to replicate past the damage and fix it later.
Regulation of Damage Control
DNA repair is regulated in mammalian cells by a sensing mechanism that detects DNA damage and activates a protein called p53. p53 is a transcriptional regulatory factor that controls the expression of some gene products that affect cell cycling, DNA replication and DNA repair. Some of the functions of p53, which are just being determined, are: stimulation of the expression of genes encoding p21 and Gaad45. Loss of p53 function can be deleterious, about 50% of all human cancers have a mutated p53 gene.
The p21 protein binds and inactivates a cell division kinase (CDK) which results in cell cycle arrest. p21 also binds and inactivates PCNA resulting in the inactivation of replication forks. The PCNA/Gaad45 complex participates in excision repair of damaged DNA.
Some examples of the diseases resulting from defects in DNA repair mechanisms.
Hereditary nonpolyposis colorectal cancer
© Dr. Noel Sturm 2019