DNA Synthesis
The discovery of the double-helical nature of DNA by Watson & Crick explained how genetic information could be duplicated and passed on to succeeding generations. The strands of the double helix can separate and serve as templates for the synthesis of daughter strands. In conservative replication the two daughter strands would go to one daughter cell and the two parental strands would go to the other daughter cell. In semiconservative replication one parental and one daughter strand would go to each of the daughter cells.
Through experimentation it was determined that DNA replicates via a semiconservative mechanism. There are three possible mechanisms that can explain DNA's semiconservative replication.
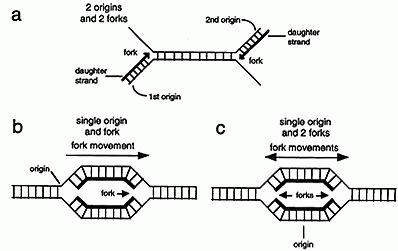
(a) DNA synthesis starts at a specific place on a chromosome called an origin. In the first mechanism one daughter strand is initiated at an origin on one parental strand and the second is initiated at another origin on the opposite parental strand. Thus only one strand grows from each origin. Some viruses use this type of mechanism.
(b) In the second mechanism replication of both strands is initiated at one origin. The site at which the two strands are replicated is called the replication fork. Since the fork moves in one direction from the origin this type of replication is called unidirectional. Some types of bacteria use this type of mechanism.
(c) In the third mechanism two replication forks are initiated at the origin and as synthesis proceeds the two forks migrate away from one another. This type of replication is called bi-directional. Most organisms, including mammals, use bi-directional replication.

Requirements for DNA Synthesis
There are four basic components required to initiate and propagate DNA synthesis. They are: substrates, template, primer and enzymes.
Substrates
Four deoxyribonucleotide triphosphates (dNTP's) are required for DNA synthesis (note the only difference between deoxyribonucleotides and ribonucleotides is the absence of an OH group at position 2' on the ribose ring). These are dATP, dGTP, dTTP and dCTP. The high energy phosphate bond between the a and b phosphates is cleaved and the deoxynucleotide monophosphate is incorporated into the new DNA strand.
Ribonucleoside triphosphates (NTP's) are also required to initiate and sustain DNA synthesis. NTP's are used in the synthesis of RNA primers and ATP is used as an energy source for some of the enzymes needed to initiate and sustain DNA synthesis at the replication fork.
Template
The nucleotide that is to be incorporated into the growing DNA chain is selected by base pairing with the template strand of the DNA. The template is the DNA strand that is copied into a complementary strand of DNA.
Primer
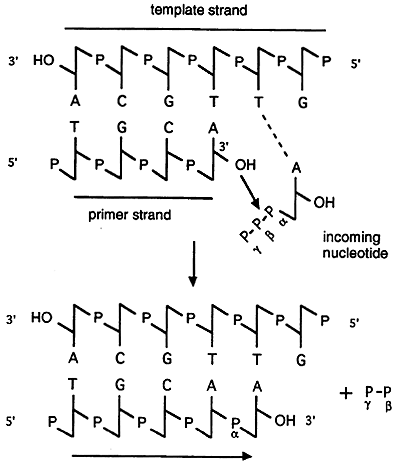
The enzyme that synthesizes DNA, DNA polymerase, can only add nucleotides to an already existing strand or primer of DNA or RNA that is base paired with the template.
Enzymes
An enzyme, DNA polymerase, is required for the covalent joining of the incoming nucleotide to the primer. To actually initiate and sustain DNA replication requires many other proteins and enzymes which assemble into a large complex called a replisome. It is thought that the DNA is spooled through the replisome and replicated as it passes through.
DNA Synthesis, 5' to 3'
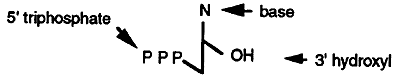
The major catalytic step of DNA synthesis is shown below. Notice that DNA synthesis always occurs in a 5' to 3' direction and that the incoming nucleotide first base pairs with the template and is then linked to the nucleotide on the primer.
DNA Synthesis is Semidiscontinuous
Since all known DNA polymerases can synthesize only in a 5' to 3' direction a problem arises in trying to replicate the two strands of DNA at the fork.
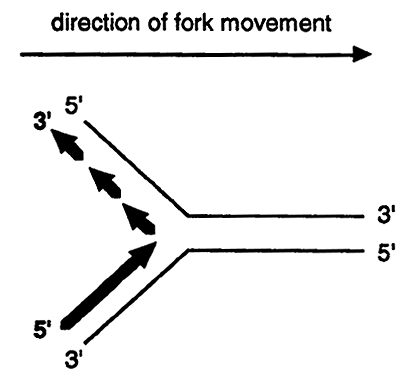
Notice that the top strand must be discontinuously replicated in short stretches thus the replication of both parental strands is a semidiscontinuous process. The strand that is continuously synthesized is called the leading strand while the strand that is discontinuously synthesized is called the lagging strand.
Leading Strand Synthesis
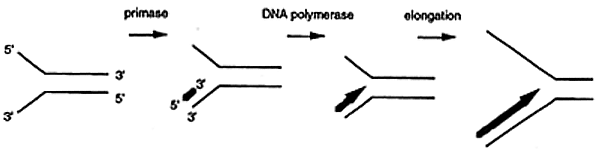
DNA synthesis requires a primer usually made of RNA. A primase synthesizes the ribonucleotide primer ranging from 4 to 12 nucleotides in length. DNA polymerase then incorporates a dNMP onto the 3' end of the primer initiating leading strand synthesis. Only one primer is required for the initiation and propagation of leading strand synthesis.
Lagging Strand Synthesis
Lagging strand synthesis is much more complex and involves five steps.
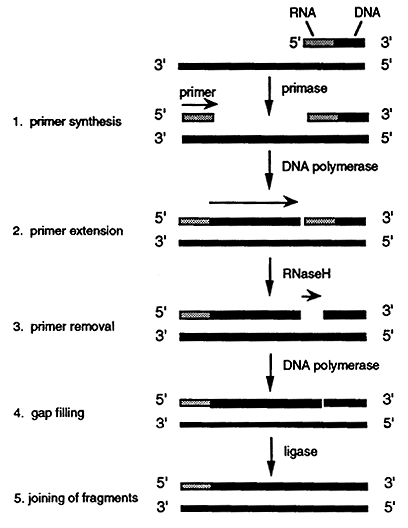
1. As the leading strand is synthesized along the lower parental strand the top parental strand becomes exposed. The strand is then recognized by a primase which synthesizes a short RNA primer.
2. DNA polymerase then incorporates a dNMP onto the 3" end of the primer and initiates lagging strand synthesis. The polymerase extends the primer for about 1,000 nucleotides until it comes in contact with the 5' end of the preceding primer. These short segments of RNA/DNA are known as Okazaki fragments.
3. When the DNA polymerase encounters the preceding primer it dissociates. The RNA is then removed by a specialized DNA polymerase or by an enzyme called RNaseH. Ribonucleotides are then excised one at a time in a 5' to 3' direction. The RNaseH leaves a phosphate group at the 5' end of the adjoining DNA segment thus leaving a gap.
4. The gap is filled by a DNA polymerase which uses an Okazaki fragment as a primer.
5. The 3' hydroxyl group on the 3' nucleotide terminus is then covalently joined, using DNA ligase, to the free 5' phosphate of the previously made lagging segment.
Structure of DNA
DNA Polymerases
There are many types of DNA polymerases which can excise, fill gaps, proofread, repair and replicate.
Other Factors Required for DNA Synthesis
Origins: Origins are unique DNA sequences that are recognized by a protein that builds the replisome. Origins have been found in bacterial, plasmid, viral, yeast and mitochondrial DNA and have recently been discovered in mammalian DNA. Specific origins are used for initiating DNA replication in humans. Most origins have a site that is recognized and bound by an origin-binding protein. When the origin-binding protein binds to the origin the A + T rich sequence becomes partially denatured allowing other replication factors known as cis-acting factors to bind and initiate DNA replication.
Origin-binding Protein: binds and partially denatures the origin DNA while binding to another enzyme called helicase.
Helicases: unwind double stranded DNA.
Single-stranded DNA Binding Protein (SSB): enhances the activity of the helicase and prevents the unwound DNA from renaturing.
Primase: synthesize the RNA primers required for initiating leading and lagging strand synthesis.
DNA Polymerase: recognizes the RNA primers and extends them in the 5' to 3' direction.
Processivity Factors: help load the polymerase onto the primer-template while anchoring the polymerase to the DNA.
Topoisomerase: removes the positive supercoils that form as the fork is unwound by the helicase.
RNaseH: removes RNA portions from Okazaki fragments.
Ligase: seals the nicks after filling in the gaps left by DNA polymerase.
Coordination of Leading and Lagging Strand Synthesis
Leading and lagging strand synthesis is thought to be coordinated at a replication fork. The two polymerases are held together by another set of proteins, tg, which are near the fork that is being unwound and simultaneously primed by helicase-primase. Both polymerases are bound by a processivity factor, b. Upon completing an Okazaki fragment the lagging strand polymerase release the b factor and dissociates from the DNA. The tg complex then loads the new b factor/primer complex onto the lagging strand polymerase which initiates a new round.....
Telomerase
Leading strand synthesis can proceed all the way to the end of a chromosome however lagging strand synthesis can not. Consequently the 3' tips of each daughter chromosome would not be replicated.
Telomerase ( also AKA telomere terminal transferase) extends the 3' ends of a chromosome by adding numerous repeats of a six base pair sequence until the 3' end of the lagging strand is long enough to be primed and extended by DNA polymerase.
Telomerase recognizes the tips of chromosomes also know as telomeres. The DNA sequences of telomeres have been determined in several organisms and consist of numerous repeats of a 6 to 8 base long sequence, [TTGGGG]n.
Telomeres have been found to progressively shorten in certain types of cells. These cells appear to lack Telomerase activity. When telomeric length shortens to a critical point the cell dies. Cells derived from rapidly proliferating tissues, such as tumors, have telomeres that are unusually long. This indicates that Telomerase activity may be necessary for the proliferation of tumor cells. Telomerase activity is found in ovarian cancer cells but not in normal ovarian tissue. Thus it may be possible to develop anti-tumor drugs that function to inhibit telomerase activity.
Chemical Inhibitors of DNA Replication
Some types of drugs function by inhibiting DNA replication.
Substrate Analogs: analogs of dNTP's which function as chain terminators can be incorporated into DNA. These analogs are usually either missing the 3' hydroxyl group or have a chemical group, other than hydroxyl, in the 3' position.
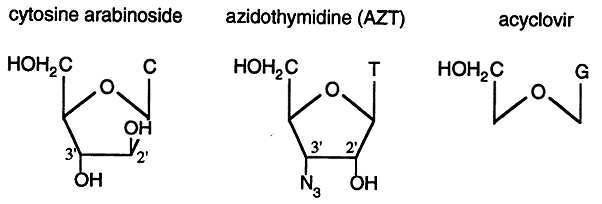
Cytosine Arabinoside: is an anticancer drug used to treat leukemia.
Azidothymidine (AZT): was used as an anti-HIV drug that, while effective in tissue culture experiments, proved to be ineffective for treating HIV in humans.
Acyclovir: is an effective anti-herpes virus drug.
Intercalating Agents: are compounds with fused aromatic ring systems that can wedge (intercalate) between the stacked base pairs of DNA. This disrupts the structure of the DNA so that the replicative enzymes have difficulty in synthesizing DNA past the "intercalated" sites. Anthracycline glycosides and Actinomycin D are intercalators used to treat a variety of cancers.
DNA Damaging Agents: a variety of compounds such as Cisplatin, cause chemical damage to DNA and are used in the treatment of cancers.
Topoisomerase Inhibitors: Nalidixic acid and Fluoroquinolones are antibiotics used to inhibit bacterial topoisomerases.
© Dr. Noel Sturm 2019